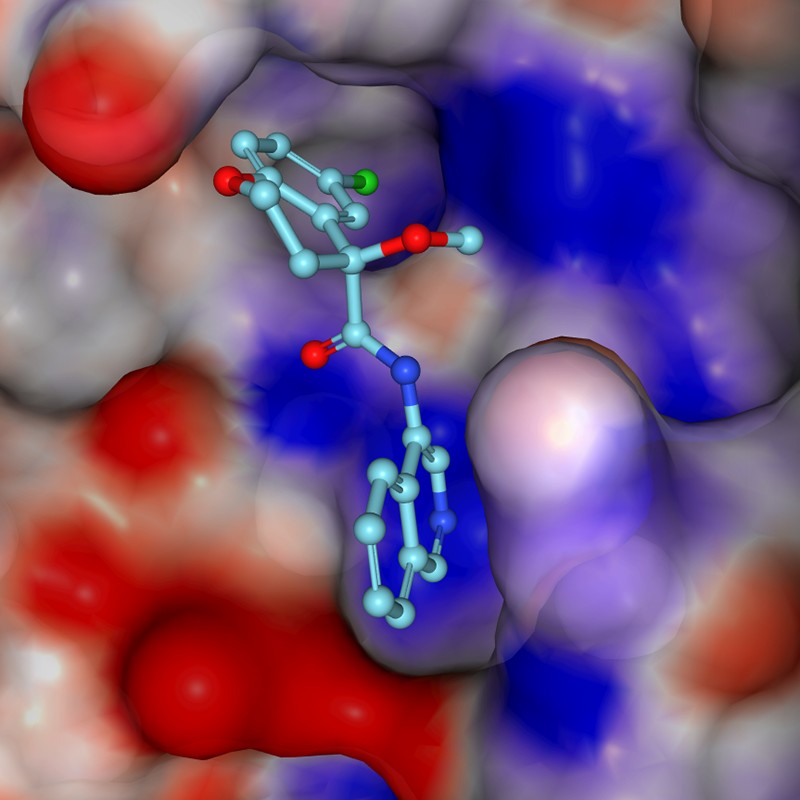
Nearly 15 months ago, a large, fast-moving and unscheduled experiment began: probing a key protein of the coronavirus SARS-CoV-2 to find chemical starting points for drug discovery. The end point was to develop pills that people could take to treat COVID-19 and related diseases.
This experiment pulled together a spontaneous, open, global, Twitter-fuelled collaboration called the COVID Moonshot. Urgency and a commitment to working openly recruited more than 150 active participants, spanning a huge range of expertise and technology across academia, biotechnology, pharmaceuticals and more, all working without claiming intellectual property. Open drug-discovery efforts are invariably super slow — ours has been an express train on tracks we have laid down as we go. It is a way of working that none of us realized was possible.
The intention for the original experiment was simply to help jump-start large drug-discovery initiatives that could draw directly on our data. In those first weeks, before the pandemic had taken hold in the United Kingdom or Israel (where the experiment started), we expected that some international effort was already in the works for countries and companies to collaborate on finding COVID-19 treatments, as was happening with vaccines.
Disappointingly, from the start of the COVID-19 fight, international funders decided to support only the development of repurposed small-molecule drugs and monoclonal antibodies to deliver treatments quickly, neglecting other approaches. The world seemed to give up on new antivirals before they even started, agreeing on a self-fulfilling prophesy that such drugs would take years to develop. Few seemed willing to contemplate such a timescale for this pandemic. Our first grant proposal was rejected, so we had to find a different way to press on.
Amazing virtual collaborations sprang up around the pandemic in many fields: bioinformaticians and phylogeneticists worked out ways to track new variants. Epidemiologists and computer modellers ran simulations. The World Health Organization activated a network of experts to vet new publications and preprints. Military personnel transported medical equipment and vaccines, and set up community testing centres.
Our COVID Moonshot is different. Rather than engaging with patients while using personal protective equipment, we work in chemistry hoods and with spectrometers, X-rays, computer models and courier companies. It’s driven by a conviction that conventional wisdom is wrong about de novo drug discovery being a job only for big pharma and peripheral to a fast-moving global outbreak: the pandemic is still here, and antiviral drugs against COVID-19 are not.
The screens
Drug-discovery efforts generally require a target, such as a protein that has an important role in disease. Promising drug compounds bind to the protein, affect its function and act safely in the body. Diamond Light Source near Oxford is the UK national synchrotron — a particle accelerator essential for modern X-ray crystallography, the go-to technique for determining 3D structures of proteins. There, one of us (F.v.D.) leads the XChem facility that uses the technique to screen for very small compounds called fragments that bind to drug targets. Although these ‘fragment hits’ bind weakly and the throughput is low compared with other techniques (screening fewer than 1,000 compounds per experiment), the 3D structures show exactly how each fragment binds. This provides powerful clues about how to create bigger, more potent molecules.
By late January 2020, scientists in China had solved the first 3D crystal structures of the SARS-CoV-2 main protease (Mpro), an essential viral enzyme, and made them public. With their guidance, a group at Diamond led by Martin Walsh generated new, high-quality crystals by mid-February — lightning fast for such work. The group also shipped Mpro protein to the Weizmann Institute of Science in Rehovot, Israel, where N.L.’s group uses mass spectrometry to quickly identify covalent fragments that attach to proteins irreversibly. This is another way to find useful starting points for drugs.
Racing to exploit the two weeks before a scheduled shutdown of the synchrotron on 6 March last year, more than a dozen scientists from the Walsh, F.v.D. and N.L. groups dropped everything to complete an XChem experiment four times the normal size1. All the data were analysed within one month, and as soon as we had the first batch of results, we posted downloadable data and a short write-up on the Diamond web page, then tweeted the link on 7 March.
The tweets
The response surprised us: almost 1,000 retweets in a week, and diverse offers for help. A.L. and M.R., two co-founders of the US–UK technology firm PostEra, got in touch to say that their machine-learning technology could propose synthetic routes to make new molecules inspired by the fragment hits. But first we needed drug-like molecules to be designed, and N.L. realized whom we could ask: medicinal chemists newly under lockdown restrictions, but full of expertise and desperate to help.
The next step was a tweet to crowdsource ideas for such molecules, declaring that we would make and test the best ones. A web page built by M.R. and his team in 48 hours enabled participants to submit machine-readable suggestions for compounds. The site made clear that contributions would have no strings attached, no intellectual property and no remuneration. We expected a few hundred submissions at most — in two weeks, we had more than 4,000, and had to work out how to test them.
The experiments
From March to May last year, we were on Zoom calls almost daily, lining up collaborators, logistics, expertise, funding, institutional support and permissions. All around us, the world was shutting down. We were trying to work out how to keep ourselves, our colleagues and our families sane, and our laboratories open.
We tapped an inexhaustible wellspring of goodwill. At the Ukrainian company Enamine, T.M. convinced management to commit to doing synthesis at cost, and to handle compound logistics. Its 650 chemists make molecules to order and have a renowned collection of building blocks for quick synthesis. By early May, new compounds were being shipped weekly from Enamine to organizations in four countries, and that work continues. Two other contract research organizations, WuXi in China and Sai Life Sciences in India, pitched in with offers of chemists and discounts.
Chris Schofield and his team at the University of Oxford, UK, together with Haim Barr and his colleagues at the Weizmann Institute, developed distinct biochemical assays that were key to cross-validating how well molecules inhibited the working Mpro enzyme. At the same time, for all compounds, the 3D mode of binding was assessed at Diamond in crystal structures. Half a dozen graduate students and postdocs suspended their own projects to coordinate, run and evaluate these assays, week after week. The work hasn’t stopped since.
By mid-April 2020, a volunteer troop of industry-based medicinal chemists, chaired by E.G., were holding weekly meetings to scrutinize submissions, review results, discuss strategies, design molecules and coordinate with synthetic chemists at Enamine. This work continues, too.
Computational chemists assembled their own team through their own network, then met weekly to work out algorithms to rank submissions. J.C. developed new ways to use Folding@home, the world’s largest crowdsourced supercomputer, which was already being used to generate models of viral proteins. It crunched ‘free energy’ calculations to predict the best binders for up to 10,000 compounds a week: 100 times more than had been attempted before.
Pharmaceutical companies develop elaborate information systems to track, store and analyse compounds and their associated data; our global effort urgently needed this, too. The informatics web platform CDD Vault donated us cloud space in its infrastructure just hours after a phone call, also arranging training and support. Many other vendors provided licences for free, and XChem’s platform for sharing 3D data, the Fragalysis cloud, had fortunately just been released. M.R. built a back-end system that sent all data live on GitHub, which is more often used as a repository for programming code.
As the pandemic unfolded, on some calls, you could hear the ambulance sirens from half a world away. The first agenda item of every meeting was a list of participants’ latest constraints — lockdowns, lab closures and home-schooling. Children made regular Zoom appearances, and at least two of us came down with COVID-19 ourselves. People pulled their weight not for glory or reward, but because there was a job that needed doing, and it was one that they could do.
To cells and live virus
By June 2020, the Zoom-based collaboration had identified sets of molecules that clearly inhibited a crucial viral protein. The next step was to test antiviral activity in living cells. These are complex experiments, requiring level-three biosafety labs certified for airborne pathogens.
A.v.D., a translational clinician, coordinated a shifting coalition of groups. One virologist friend and colleague lived a 10-minute walk away, and they planned experiments on lockdown evening strolls. Other virology groups responded to our tweet for help, and offered a variety of assays. Compounds were shipped, early results trickled in and some compounds unambiguously stalled the virus. These initial successes were crucial, both scientifically and for morale.
Researchers at the Israel Institute for Biological Research near Rehovot agreed to run a single test plate once we had molecules that were sufficiently potent. When that test showed signs of drug-like activity, they worked out how to conduct regular measurements, filling a crucial gap in our testing cascade.
By September, we had reached a milestone with a chemical series that instilled confidence: the compounds inhibited enzymes at submicromolar concentrations, and blocked viral activity at single-digit micromolar concentrations.
The slog
Since then, for the past nine months, the project has entered familiar territory in medicinal chemistry: we have been tweaking and testing compound designs, and optimizing early lead molecules so that they behave like drugs — entering the blood and staying there without being toxic. Potency against the Mpro enzyme has improved 100-fold, as has antiviral activity, and we are honing compounds’ solubility and rate of metabolism by the liver.
Above all, we can start predicting that these molecules will be straightforward to synthesize and will work as pills that are suitable for vaccine-hesitant or immunocompromised individuals, health-care workers and others in risky situations who could take them prophylactically. Furthermore, we expect them to work against vaccine-resistant variants: whereas vaccines target the spike protein on the virus capsule, our compounds target a conserved part of the virus machinery that works inside cells.
We’ve also had to deal with rejected grant proposals to advance antiviral drugs. Still, as vaccines have showed their dramatic successes, further variants have arrived and funders have begun calling urgently for antivirals and looking at how projects might be accelerated. In April this year, 16 months after the outbreak of SARS-CoV-2 in Wuhan, China, the United Kingdom finally launched a task force focusing on antivirals2.
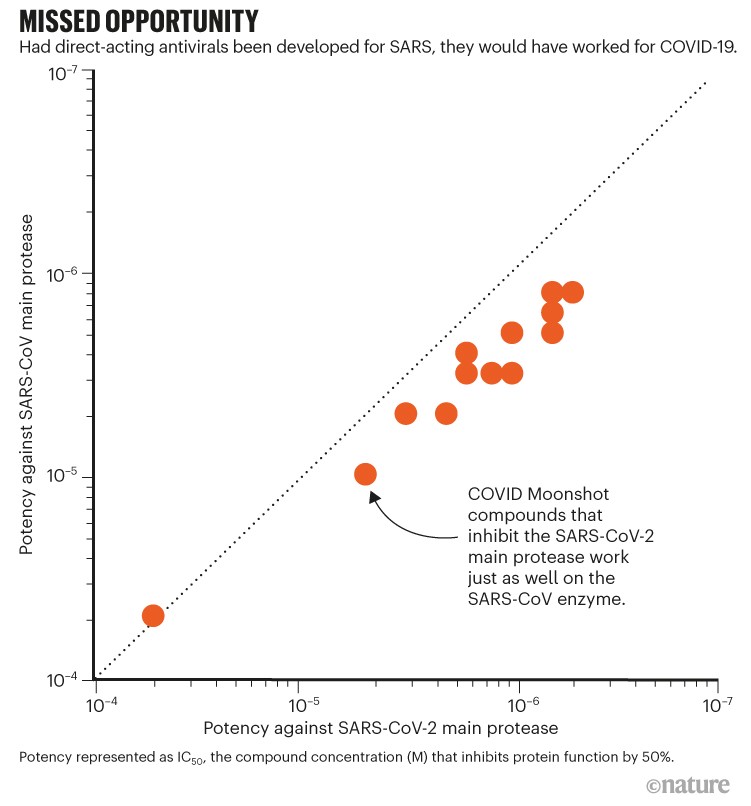
Pfizer’s March announcement of early clinical trials for its antiviral pill is confirmation that an accelerated approach can work, and that we should persevere. Our molecules also inhibit proteins of the coronavirus that causes severe acute respiratory syndrome (SARS; see ‘Missed opportunity’): had drug discovery persevered during the SARS epidemic in 2003, antiviral drugs would have been available when this pandemic hit. Above all, it has become much clearer how an antiviral would be most effective: the treatment must be readily available to everybody, long before they are hospitalized. Accordingly, we have been able to develop a clear plan for how to proceed, and the resources required.
We are approaching the capital-intensive, highly regulated phases of animal studies, producing kilograms of substance for clinical trials and, beyond that, worldwide manufacture and distribution of billions of pills. Our initial goal of delivering a drug straight from the discovery pipeline, free from patents and available for anyone to manufacture, cannot offer investors any conventional return on investment. Yet COVID-19 is not conventional, and vaccines have elevated the normally arcane question of intellectual property into a major political concern. Perhaps the COVID Moonshot can also shape how open drug discovery reaches patients.
The moral
So, what has made our approach work? Presumably, the fact that the mission was clear, even if distant, and the ethos was unambiguous and clearly signposted3,4. Initially, a few of us, fuelled by the urgency of the moment, acted on a conviction that our various combined technologies would accelerate drug discovery. We were soon joined by many people who did the hard work because they felt it was the right thing to do.
Also crucial was the existing large ecosystem of expertise and biopharma supply chains, coupled with new capabilities driven by long-term strategic investments in national infrastructure and research institutes. Tools for online collaboration have reached a critical mass, both general ones (such as Zoom or Google Docs) and those specific to drug discovery (in our case, CDD Vault). Serendipitously, for the segments of our project that had the most collaborators — such as submitting ideas for molecules — the requested contributions broke into discrete, doable tasks that easily accommodated each contributor’s availability and know-how.
The project self-selected a team of reflexively collaborative people, with no big egos. So far, we have avoided bureaucracy — no one claims to be the head of the COVID Moonshot. We retained momentum with collective trust, combined with sufficiently diverse expertise and perspectives, which allowed us to rapidly reach and implement strategic decisions. Reassuringly, people seemed to leave the collaboration only once their part of the project had been completed.
Perhaps the most surprising asset was that we did not have time to plan much at all — if we had, we’d have been paralysed. It seems you just have to get started and set deadlines for when to move on. Even now, we are astonished at how quickly this infrastructure self-assembled, just by scientists unabashedly asking for help from colleagues, distant connections or vendors. With so clear a goal, so obvious a need and the complete absence of contracts, people across the world stepped up.
Competing Interests
J.C. is a current member of the Scientific Advisory Board of OpenEye Scientific Software, Redesign Science, and Interline Therapeutics, and has equity interests in Redesign Science and Interline Therapeutics. His group receives or has received funding from multiple sources, including the National Institutes of Health, the National Science Foundation, the Parker Institute for Cancer Immunotherapy, Relay Therapeutics, Entasis Therapeutics, Silicon Therapeutics, EMD Serono (Merck KGaA), AstraZeneca, Vir Biotechnology, Bayer, XtalPi, Foresite Laboratories, the Molecular Science. F.v.D.’s group receives and has received funding from the Structural Genomics Consortium and the SABS Centre for Doctoral Training, which are open science public-private partnerships that have collectively involved over 30 companies over the last 18 years. E.G. is a Director of MedChemica Ltd, MedChemica Consultancy Ltd and MedChemica Holdings Ltd; he is is a shareholder in MedChemica Holdings Ltd and AstraZeneca plc. A.L. is the Chief Scientific Officer and a shareholder of PostEra Inc. His group at the University of Cambridge receives funding from multiple sources, including Pfizer, AstraZeneca, the Engineering and Physical Sciences Research Council, The Royal Society, and the Winton Programme for the Physics of Sustainability. N.L. is a member of the scientific advisory board of Monte Rosa Therapeutics, Totus Medicines and MetaboMed. His group lab has received funding from Pfizer, and Teva Pharmaceuticals. M.R. is the Chief Technology Officer and a shareholder of PostEra Inc.
F.v.D. and J.C. have been actively involved in, and vocal about, open science initiatives and partnerships for most of their research careers. E.G. undertakes software design and research and consultancy work in the pharmaceutical industry. In the anti-infective area, he consults for Bugworks Research Inc. which is focused on discovering new antibiotics for to treat resistant bacterial infections.